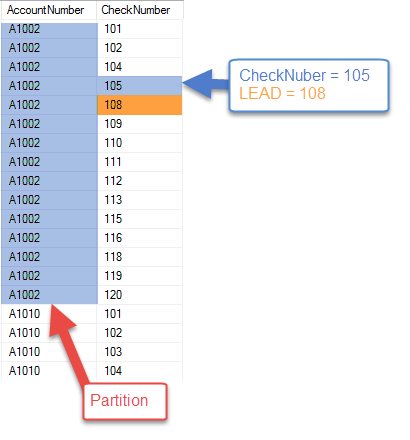
How to Interpret BLAST Results. A short summary of the quality metrics… | by Grace Reed | The Computational Biology Magazine | Medium
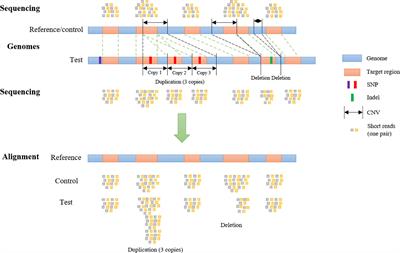
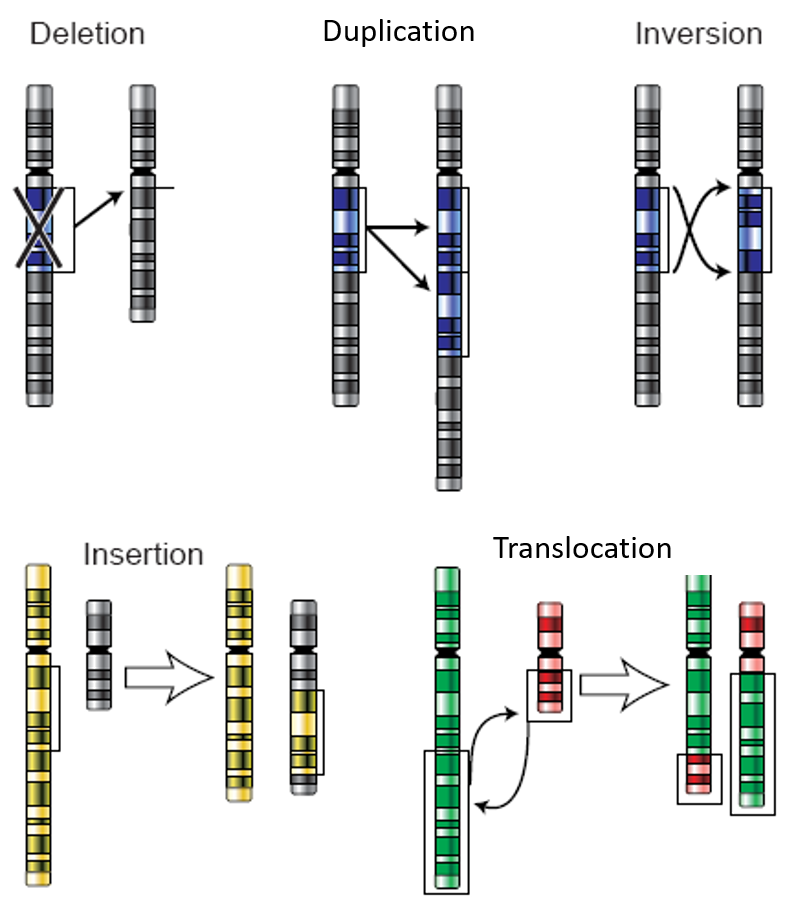
Frontiers | SECNVs: A Simulator of Copy Number Variants and Whole-Exome Sequences From Reference Genomes
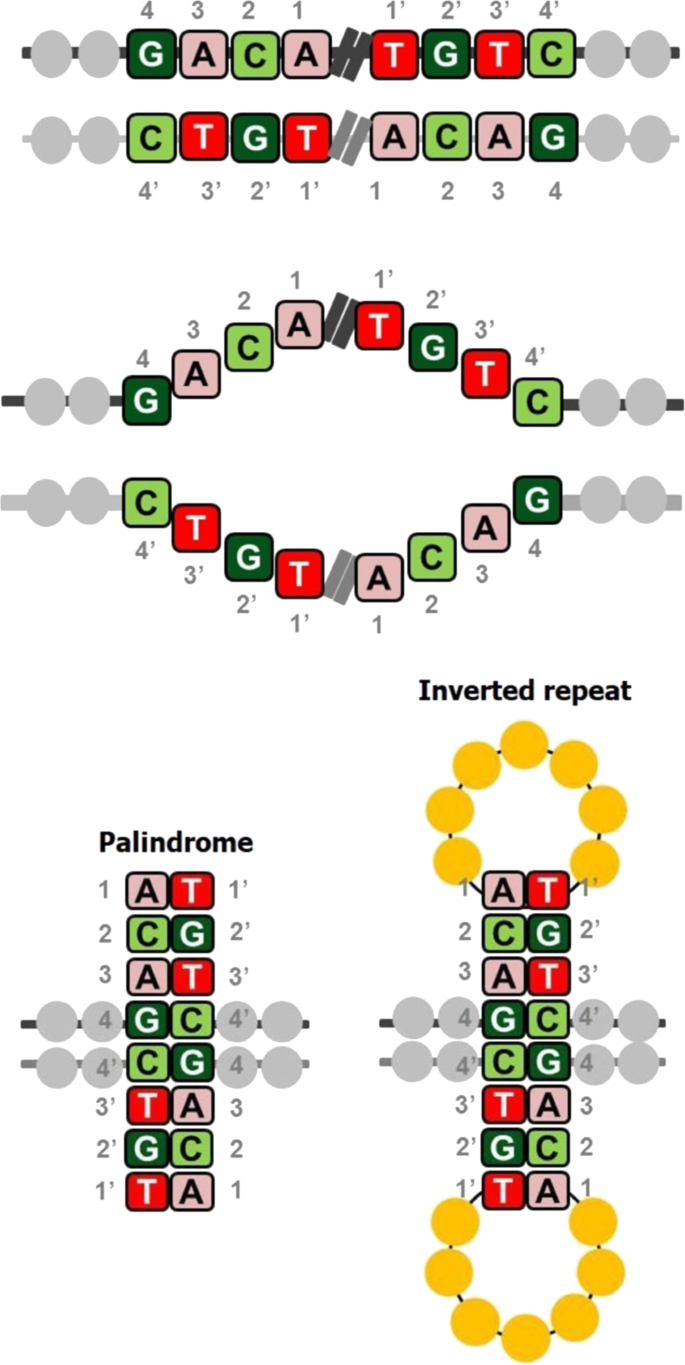
A reference catalog of DNA palindromes in the human genome and their variations in 1000 Genomes | Human Genome Variation
Calculate the Number of Gap Positions in a Sequence Alignment (and Coverage) · Issue #491 · biojava/biojava · GitHub

Gap Analysis: How to Bridge the Gap Between Performance and Potential | Process Street | Checklist, Workflow and SOP Software

Position-specific gap penalties. An alignment of two profiles X and Y.... | Download Scientific Diagram
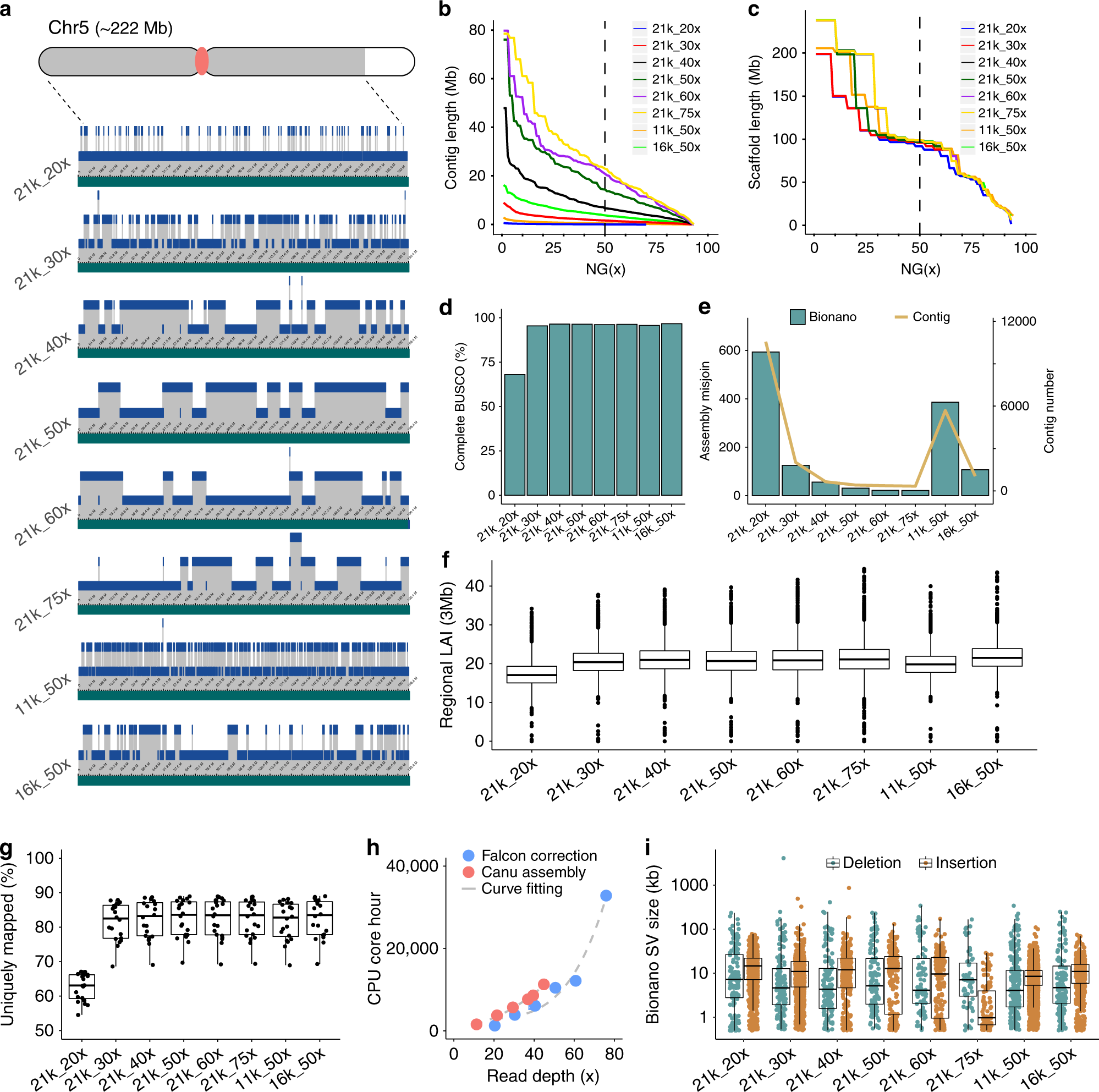
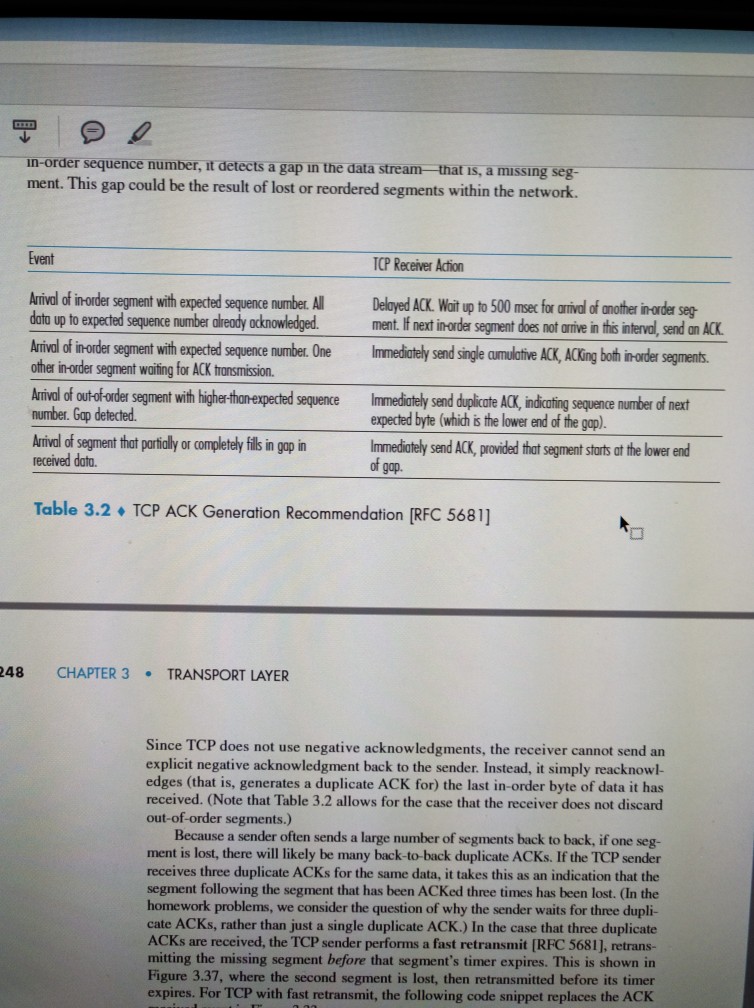
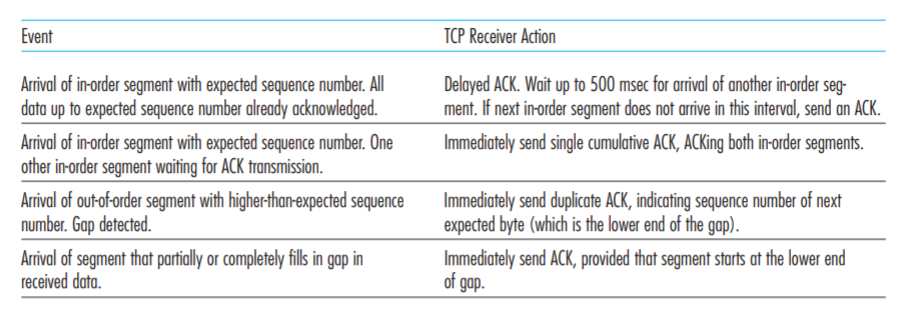
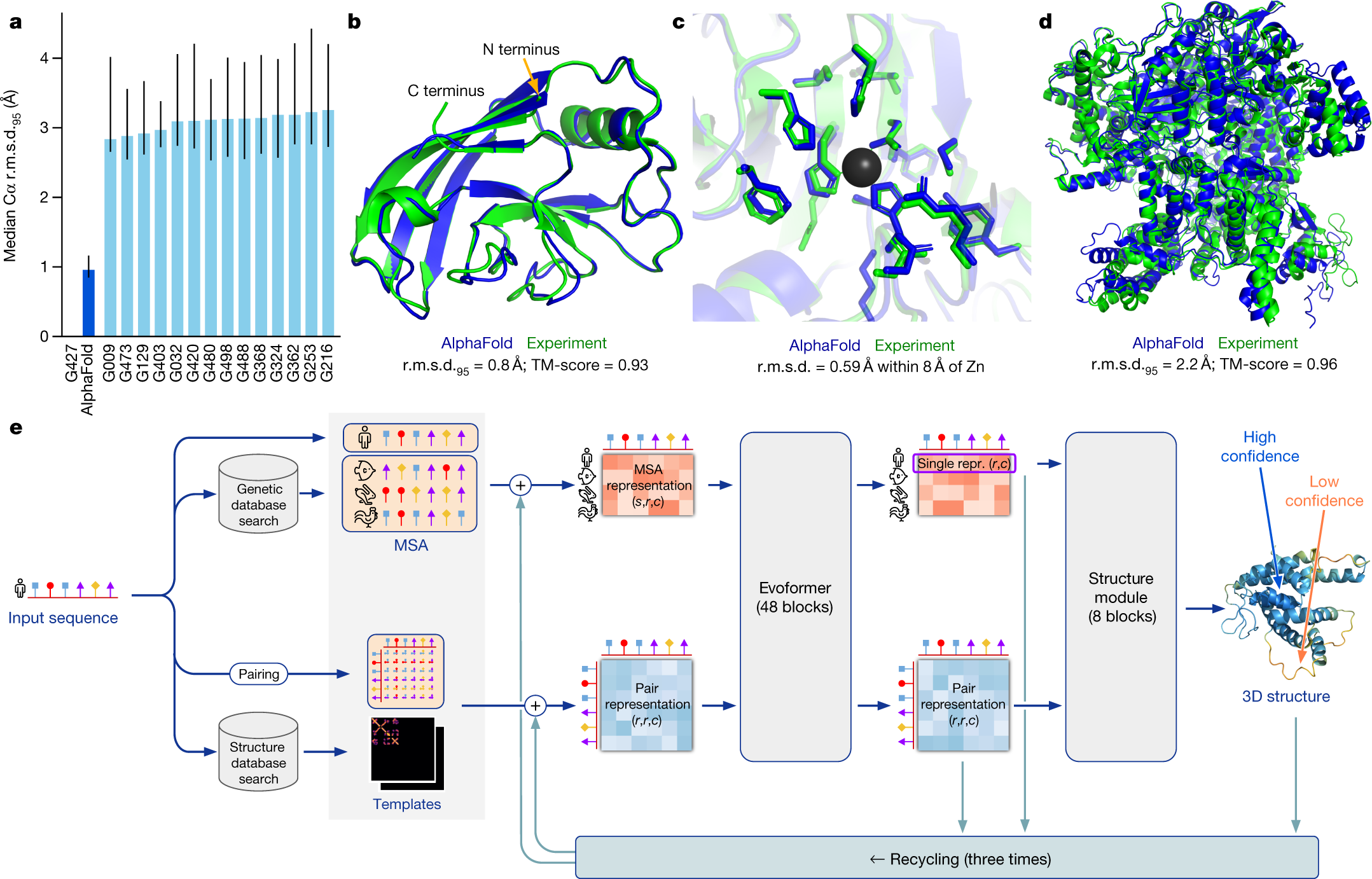
Effect of sequence depth and length in long-read assembly of the maize inbred NC358 | Nature Communications